Learn Chip-Seq & Scatac-Seq Data Analysis Using Linux And R
Published 7/2025
Created by Rafiq Ur Rehman
MP4 | Video: h264, 1280x720 | Audio: AAC, 44.1 KHz, 2 Ch
Level: Beginner | Genre: eLearning | Language: English | Duration: 24 Lectures ( 5h 2m ) | Size: 2.1 GB
Understand the principles and applications of ChIP-Seq and ATAC-Seq in studying protein-DNA interactions and chromatin accessibility.
Set up and navigate a Linux environment for bioinformatics, including using WSL (Windows Subsystem for Linux) and installing necessary tools.
Download, compress, and perform quality control of raw ChIP-Seq and ATAC-Seq data using command-line bioinformatics tools.
Index reference genomes and align sequencing reads using BWA or Bowtie2, and manage alignment files (SAM/BAM).
Perform peak calling with MACS2 and interpret the significance of enriched regions in ChIP-Seq data.
Annotate peaks and identify motifs using HOMER to gain functional insights into regulatory elements.
Learn the theory and workflow of single-cell ATAC-Seq, including its experimental setup and data structure.
Analyze scATAC-Seq data using R, Signac, and Seurat, including filtering cells, dimensionality reduction, and clustering.
Compare chromatin accessibility between different samples or conditions, and visualize the results using R plots.
Build complete ChIP-Seq and scATAC-Seq analysis pipelines that can be adapted to various research and publication needs.
Requirements
No prior experience in bioinformatics is required: this course starts from the basics and builds your skills step-by-step.
A basic understanding of biology or genomics is helpful, but not mandatory.
A laptop or desktop with at least 8 GB of RAM is recommended for smooth performance.
Description
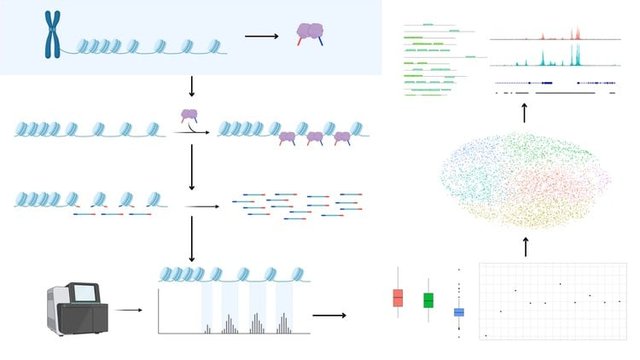
Are you ready to dive into the world of gene regulation and chromatin accessibility using cutting-edge bioinformatics tools? This course, "Master ChIP-Seq and ATAC-Seq Analysis Using Linux and R," is your gateway to understanding and performing real-world epigenomics data analysis from scratch.Designed for students, researchers, and professionals in bioinformatics, biotechnology, and genomics, this hands-on course will take you on a comprehensive journey through ChIP-Seq (Chromatin Immunoprecipitation Sequencing) and ATAC-Seq (Assay for Transposase-Accessible Chromatin using sequencing) data analysis using powerful open-source tools.You'll start by understanding the theoretical concepts behind ChIP-Seq and ATAC-Seq technologies, including how they're used to investigate protein-DNA interactions and chromatin accessibility in cells. These insights are essential in fields like cancer research, developmental biology, immunology, and beyond.What You'll LearnThe complete bioinformatics pipeline for ChIP-Seq and ATAC-SeqSetting up a Linux environment (even on Windows) for analysisRunning quality checks and trimming raw sequencing readsAligning reads to a reference genome and handling alignment filesPerforming peak calling with MACS2Annotating peaks and discovering DNA motifs using HOMERUnderstanding how single-cell ATAC-Seq worksAnalyzing scATAC-Seq datasets in R using the Signac and Seurat packagesVisualizing chromatin accessibility and comparing multiple samplesHands-On Linux and R TrainingNo prior experience with Linux or R? No problem. This course includes a full section on Linux for bioinformatics, where you'll learn to navigate the file system, run basic commands, and handle biological data on the command line.When working with single-cell ATAC-Seq data, we'll shift to R-a powerful language for statistical analysis and visualization. You'll get comfortable using RStudio, installing relevant packages, and working with real datasets from publicly available repositories like 10X Genomics and ENCODE.Real Data. Real Tools. Real Skills.Throughout this course, you'll work with real ChIP-Seq and ATAC-Seq data to apply what you learn immediately. By the end of the course, you'll be able to build your own analysis pipelines, interpret results, and even contribute to publications or research projects.We cover essential tools used by professionals worldwide, including:FastQC for quality controlBWA or Bowtie2 for alignmentMACS2 for peak callingHOMER for peak annotation and motif discoverySignac and Seurat for scATAC-Seq analysis in RWho This Course Is ForStudents in bioinformatics, computational biology, or life sciencesResearchers analyzing epigenetic or chromatin dataProfessionals looking to upskill in genome data analysisAnyone curious about sequencing data and open-source toolsNo Prior Experience RequiredWhether you're a beginner or someone with basic knowledge of sequencing technologies, this course is structured to build your skills progressively from theory to practice.Join now and gain the confidence to analyze and interpret ChIP-Seq and ATAC-Seq data like a pro. Let's uncover the regulatory code of the genome one peak at a time.
Who this course is for
Students and graduates in bioinformatics, biotechnology, molecular biology, genetics, or related life science fields who want to learn real-world data analysis skills.
Researchers and lab scientists looking to analyze ChIP-Seq or ATAC-Seq data on their own using open-source tools.
Bioinformatics beginners who want to get comfortable with Linux and R in the context of sequencing data analysis.
Data analysts and programmers transitioning into genomics or looking to apply coding skills in biological datasets.
Educators and PhD candidates who want to build a reproducible pipeline for ChIP-Seq or single-cell ATAC-Seq analysis.
Anyone curious about gene regulation, epigenomics, and single-cell technologies, and eager to explore these fields with hands-on projects.